Used to 'transfer' information from nodes to other nodes across specific predicates, either in an outward direction (along the edge direction) or inward (against the edge direction). Returns a node-property column; intended to be used with mutate() on nodes.
transfer(colname = NULL, over, direction = "out")Arguments
Value
Vector or list, with one entry per node.
Details
The return value will be either a list, or if the result would be a list with all length-1 or length-0 elements, a vector with 0-length elements replaced by NA. Practically, this results in a list when necessary and a vector otherwise.
See also
Examples
library(dplyr)
library(tidygraph)
data(eds_marfan_kg)
eds_marfan_kg |>
fetch_nodes("biolink:Gene" %in_list% category, limit = 5) |>
expand(categories = "biolink:Disease") |>
activate(nodes) |>
mutate(caused_by_genes = transfer(name,
over = "biolink:causes",
direction = "out"
)) |>
mutate(causes_diseases = transfer(name,
over = "biolink:causes",
direction = "in"
)) |>
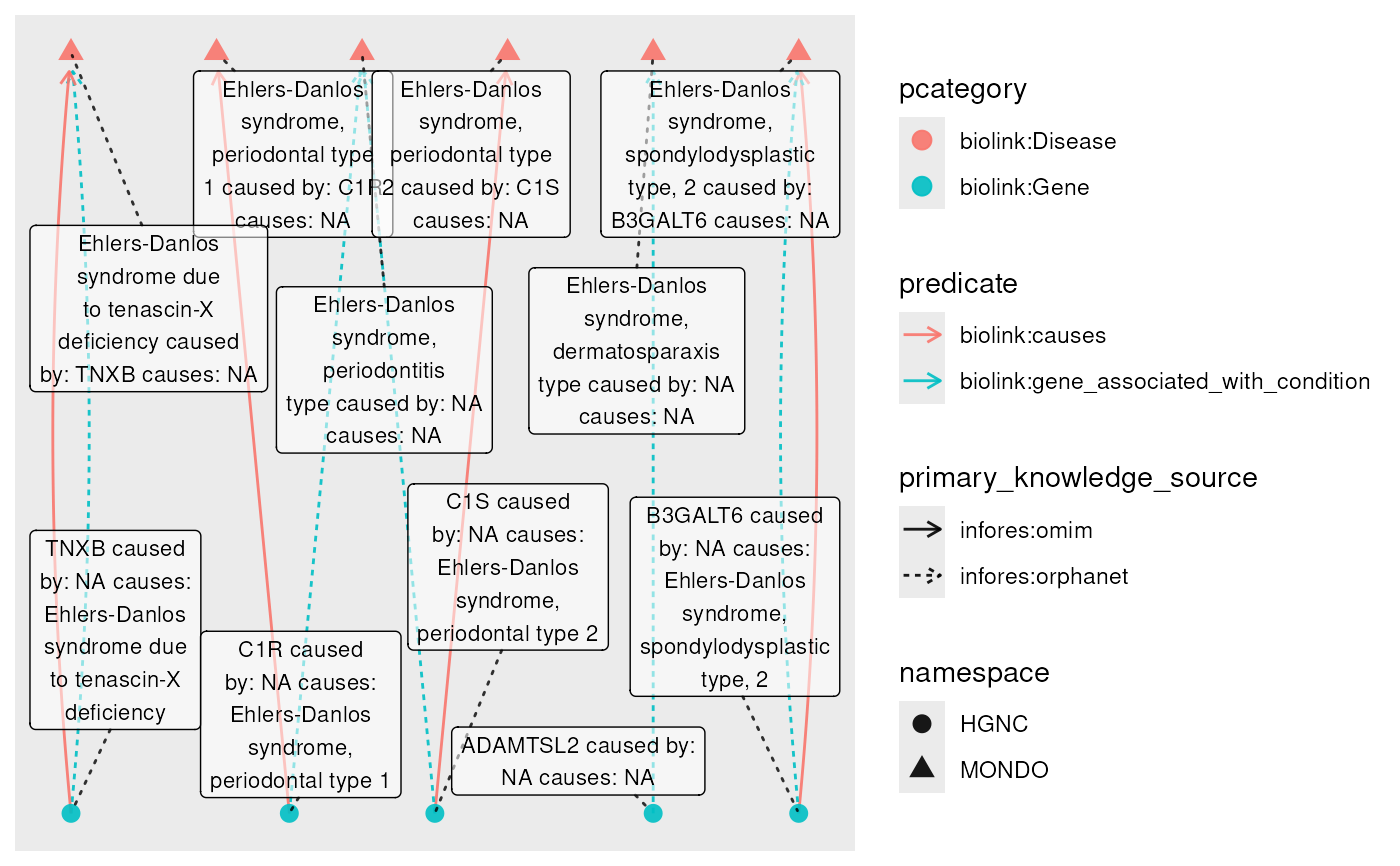
plot(
node_label = paste(
name,
" caused by: ", caused_by_genes,
" causes: ", causes_diseases
),
label_size = 3
)
#> Using "sugiyama" as default layout