Compute transitive reduction over a predicate.
Source:R/transitive_reduction.R
transitive_reduction.RdComputes the transitive reduction of a graph, treating the specified predicate as transitive.
transitive_reduction(g, predicate = "biolink:subclass_of")Value
Graph with transitive edges added.
See also
Examples
library(dplyr)
library(tidygraph)
data(eds_marfan_kg)
g <- eds_marfan_kg |>
fetch_nodes(name == "Tall stature") |>
expand_n(predicates = "biolink:subclass_of", direction = "out", n = 3) |>
bind_edges(data.frame(
from = 2,
to = 9,
predicate = "biolink_subclass_of",
primary_knowledge_source = "hand_annotated"
))
#> Initial graph size:1nodes ||0edges
#> Expanding graph: iteration 1/3
#> Graph size:3nodes ||2edges
#> Expanding graph: iteration 2/3
#> Graph size:7nodes ||7edges
#> Expanding graph: iteration 3/3
#> Graph size:13nodes ||15edges
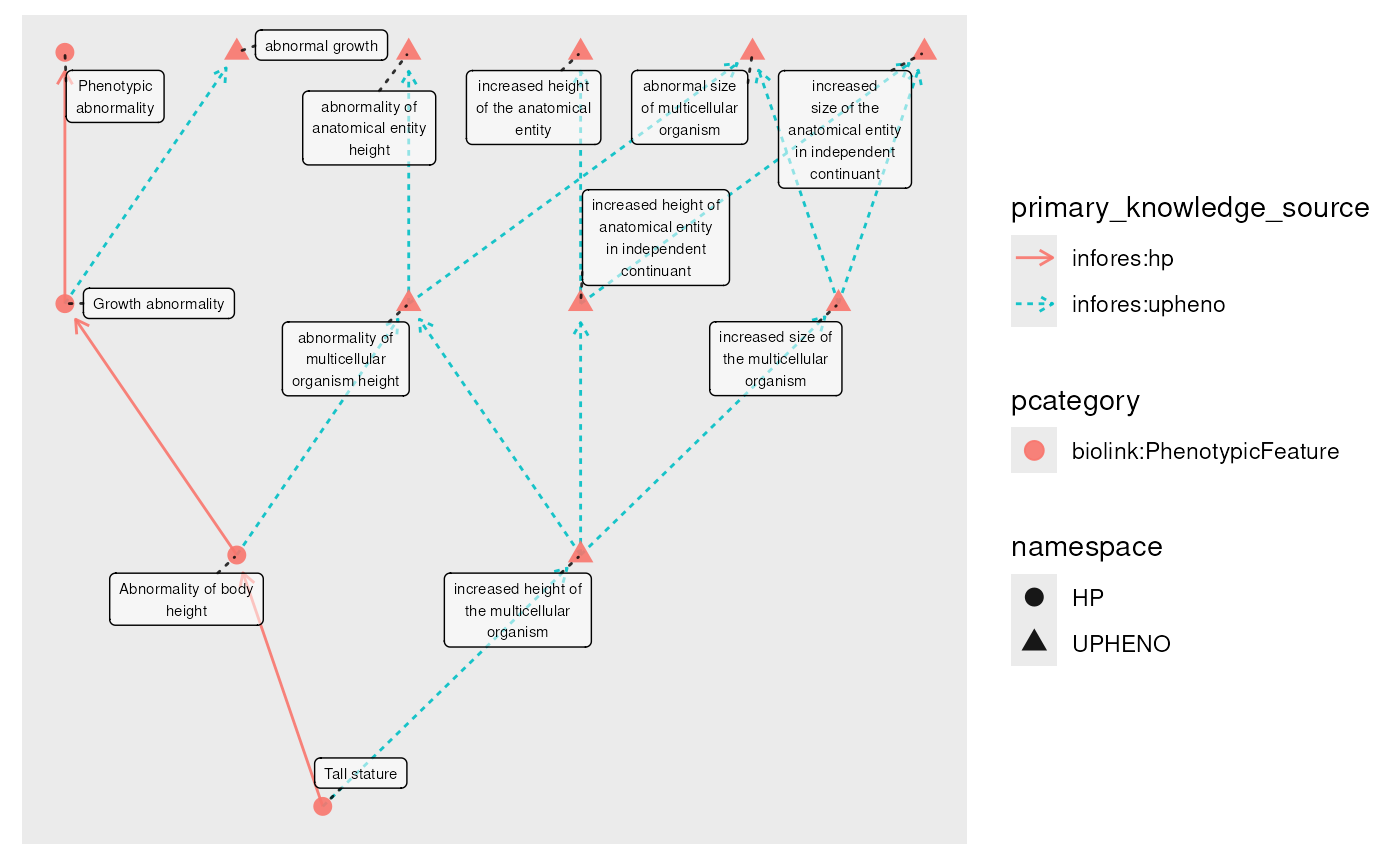
plot(g, edge_color = primary_knowledge_source)
#> Using "sugiyama" as default layout
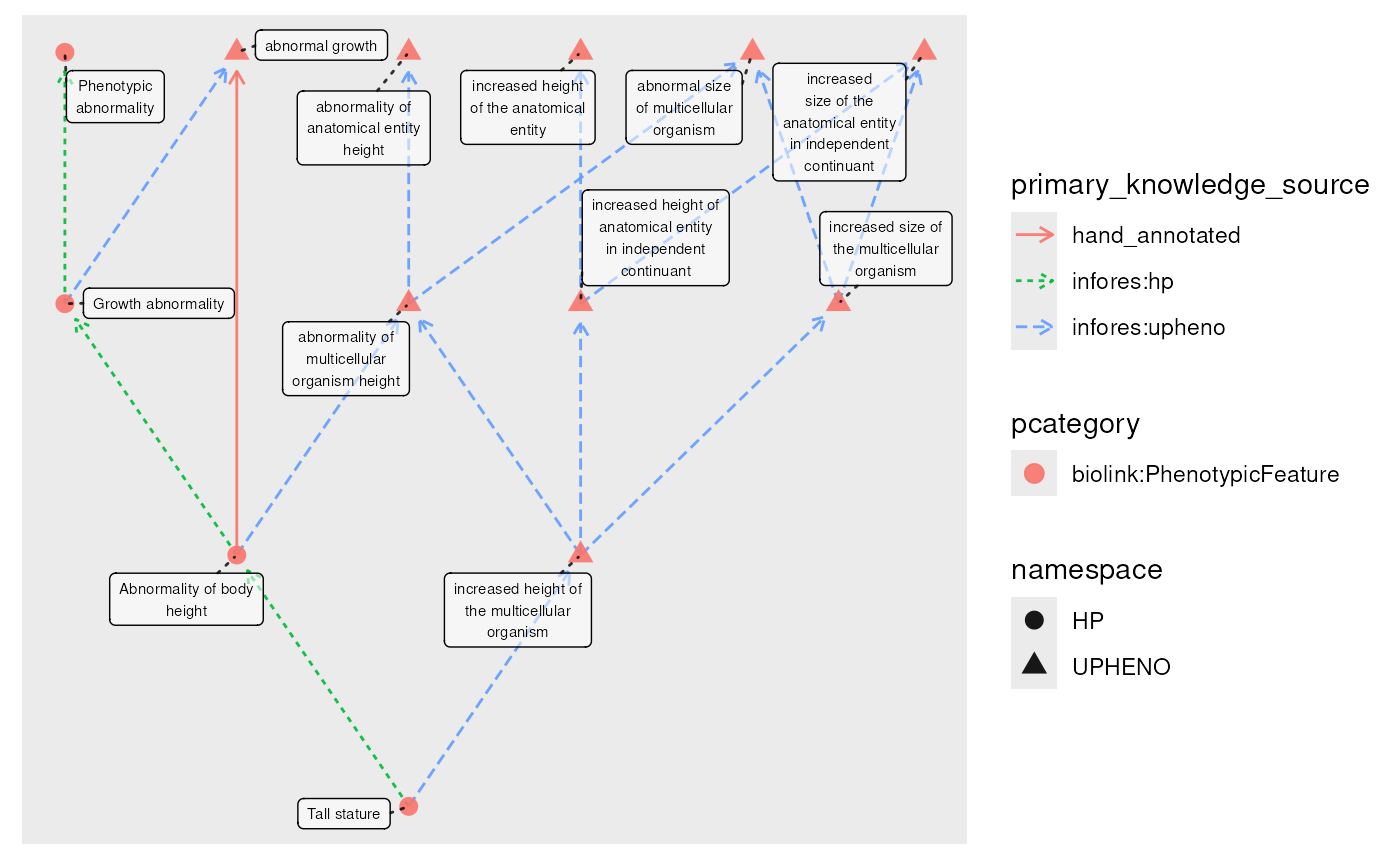 g_closed <- g |>
transitive_closure(predicate = "biolink:subclass_of")
plot(g_closed, edge_color = primary_knowledge_source)
#> Using "sugiyama" as default layout
g_closed <- g |>
transitive_closure(predicate = "biolink:subclass_of")
plot(g_closed, edge_color = primary_knowledge_source)
#> Using "sugiyama" as default layout
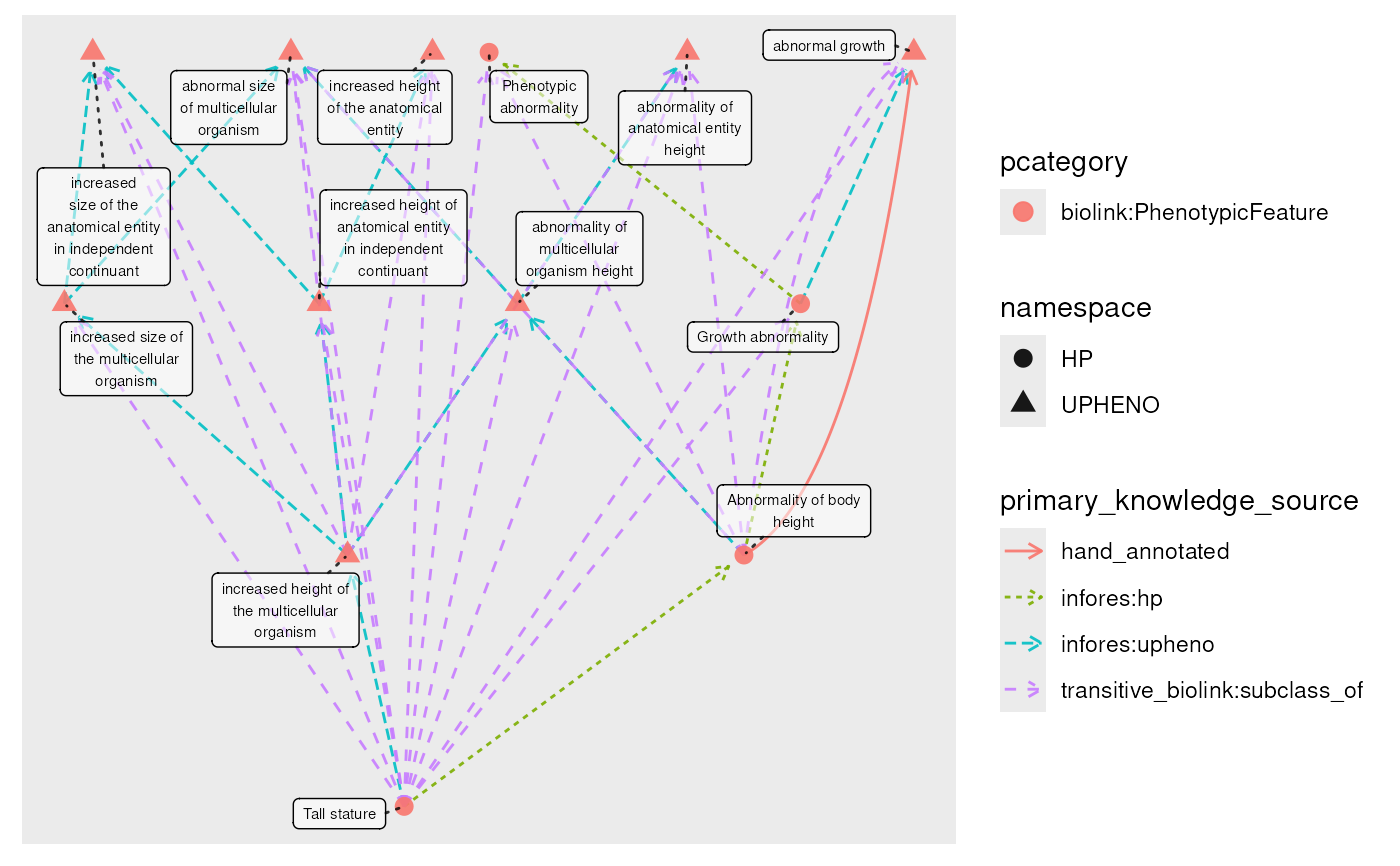 g_reduced <- g_closed |>
transitive_reduction()
plot(g_reduced, edge_color = primary_knowledge_source)
#> Using "sugiyama" as default layout
g_reduced <- g_closed |>
transitive_reduction()
plot(g_reduced, edge_color = primary_knowledge_source)
#> Using "sugiyama" as default layout