Compare genotype and phenotype classes
In this section, we show how to test the association between genotype and phenotype classes. We assume a cohort was preprocessed following the Input data section, and we use classifiers described in the Partitioning to assign each cohort member into a class along the genotype and phenotype axes. We use Fisher exact test (FET) to test for differences between the classes and we apply multiple testing correction to mitigate finding significant associations by chance.
Fisher exact test
The Fisher exact test calculates the exact probability value for the relationship between two dichotomous variables. In our implementation, the two dichotomous variables are the genotype and the phenotype. For instance, the individuals of the cohort may be divided according to whether or not they have a nonsense variant and according to whether or not they have Strabismus (HP:0000486).
The results of FET are expressed in terms of an exact probability (P-value), varying within 0 and 1. Two groups are considered statistically significant if the P-value is less than the chosen significance level (usually \(\alpha = 0.05\)).
The following graphic shows an example contingency table that is used to conduct a Fisher exact test. We are comparing the frequency of strabismus in individuals with missense and nonsense variants:
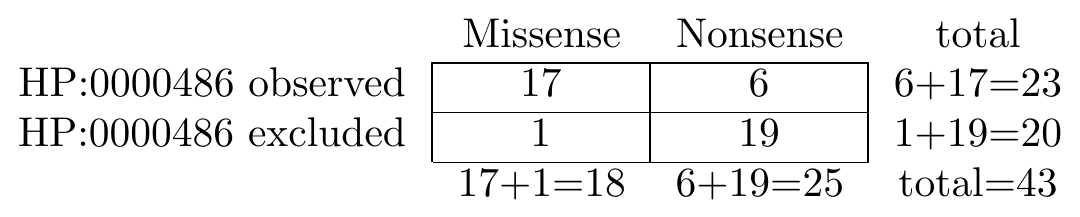
To perform the corresponding test in Python, we would use the following code.
>>> import scipy.stats as stats
>>> contingency_table = [
... # Missense, Nonsense
... [17, 6 ], # Strabismus observed
... [1, 19 ], # Strabismus excluded
... ]
>>> oddsratio, p_value = stats.fisher_exact(contingency_table)
>>> float(oddsratio)
53.833333333333336
>>> float(p_value)
5.432292015291845e-06
The p_value evaluates to 5.432292015291845e-06, meaning there is a significant difference between the groups.
The Fisher exact test evaluates whether the observed frequencies in a contingency table significantly deviate from the frequencies we would expect if there were no association between the variables. We want to test whether the frequency of HP:0000486` is significantly higher or lower in one genotype class compared to what would be expected if there were no association. Note that by default, the two-tailed Fisher exact test is performed, meaning we have no hypothesis as to whether there is a higher or lower frequency in one of the genotype classes.
However, we are typically interested in testing the associations between the genotype and multiple phenotypic features at once. GPSEA takes advantage of the HPO structure and simplifies the testing for all HPO terms encoded in the cohort.
Example analysis
Let’s illustrate this in a real-life example of the analysis of the association between frameshift variants in TBX5 gene and congenital heart defects in the dataset of 156 individuals with mutations in TBX5 whose signs and symptoms were encoded into HPO terms, stored as phenopackets of the GA4GH Phenopacket Schema, and deposited in Phenopacket Store.
Note
The shorter version of the same analysis has been presented in the Tutorial.
Load cohort
For the purpose of this analysis, we will load the Cohort
from a JSON file.
The cohort was prepared from phenopackets as described in Create a cohort section,
and then serialized as a JSON file following the instructions in Persist the cohort for later section.
>>> import json
>>> from gpsea.io import GpseaJSONDecoder
>>> fpath_cohort_json = 'docs/cohort-data/TBX5.0.1.20.json'
>>> with open(fpath_cohort_json) as fh:
... cohort = json.load(fh, cls=GpseaJSONDecoder)
>>> len(cohort)
156
Configure analysis
We want to test the association between frameshift TBX5 variants and phenotypic abnormalities. GPSEA exposes a flexible classifier API that lets us create genotype and phenotype classifiers to assign the cohort members into genotype and phenotype categories based on the variants and the HPO terms. We need to create one genotype classifier and one or more phenotype classifiers.
Genotype classifier
We want to separate the patients into two classes: a class with a frameshift variant and a class without a frameshift variant (i.e. any other heterozygous variant). We will use the MANE transcript for the analysis:
>>> tx_id = 'NM_181486.4'
Building a genotype classifier is a two step process.
First, we create a VariantPredicate
to test if the variant is predicted to lead to a frameshift in NM_181486.4:
>>> from gpsea.model import VariantEffect
>>> from gpsea.analysis.predicate import variant_effect
>>> is_frameshift = variant_effect(VariantEffect.FRAMESHIFT_VARIANT, tx_id)
>>> is_frameshift.description
'FRAMESHIFT_VARIANT on NM_181486.4'
Note
The gpsea.analysis.predicate documentation lists all available variant predicates
and Variant Predicates exemplifies their usage.
To build a genotype classifier, we wrap is_frameshift
in a Monoallelic classifier (monoallelic_classifier),
to classify each TBX5 cohort member either as an individual with one frameshift allele (Frameshift)
or as an individual with one non-frameshift allele (Other):
>>> from gpsea.analysis.clf import monoallelic_classifier
>>> gt_clf = monoallelic_classifier(
... a_predicate=is_frameshift,
... a_label="Frameshift",
... b_label="Other",
... )
>>> gt_clf.class_labels
('Frameshift', 'Other')
Note
See the Genotype classifiers for other genotype classifier examples.
In the subsequent analysis, gt_clf assigns an individual into a genotype class. Note, any individual with \(0\) or \(\ge 2\) alleles will be omitted from the analysis.
Phenotype classifiers
We recommend testing the genotype phenotype association for all HPO terms that annotate the cohort members,
while taking advantage of the HPO graph structure and of the True path rule.
We will use the prepare_classifiers_for_terms_of_interest()
utility function to generate phenotype classifiers for all HPO terms.
The function needs HPO to prepare classifiers, hence we need to load HPO:
>>> import hpotk
>>> store = hpotk.configure_ontology_store()
>>> hpo = store.load_minimal_hpo(release='v2024-07-01')
and then we can create the classifiers
>>> from gpsea.analysis.clf import prepare_classifiers_for_terms_of_interest
>>> pheno_clfs = prepare_classifiers_for_terms_of_interest(
... cohort=cohort,
... hpo=hpo,
... )
>>> len(pheno_clfs)
369
The function finds 369 HPO terms that annotate at least one individual, including the indirect annotations whose presence is implied by the True path rule.
Statistical analysis
We will use Fisher exact test to test the association between genotype and phenotype classes, as described previously.
In the case of this cohort, we can test association between having a frameshift variant and one of 369 HPO terms. However, testing multiple hypotheses on the same dataset increases the risk of finding a significant association solely by chance. GPSEA uses a two-pronged strategy to reduce the number of tests and, therefore, mitigate this risk: a phenotype multiple testing (MT) filter and multiple testing correction (MTC).
Phenotype MT filter selects a (sub)set of HPO terms for testing,
for instance only the user-selected terms (see SpecifiedTermsMtcFilter)
or the terms selected by IfHpoFilter.
MTC then adjusts the nominal p values for the increased risk of false positive G/P associations. The available MTC procedures are listed in the Multiple-testing correction procedures section.
We must choose a phenotype MT filter as well as a MTC procedure to perform genotype-phenotype analysis.
Default analysis
We recommend using Independent filtering for HPO (IfHpoFilter)
and Benjamini-Hochberg MT correction.
The default analysis can be configured with configure_hpo_term_analysis() convenience method.
>>> from gpsea.analysis.pcats import configure_hpo_term_analysis
>>> analysis = configure_hpo_term_analysis(hpo)
At this point, the analysis configured to test
a cohort for G/P associations.
Custom analysis
If the default selection of phenotype MT filter and multiple testing correction is not an option, we can configure the analysis manually.
First, we choose a phenotype MT filter (e.g. IfHpoFilter):
>>> from gpsea.analysis.mtc_filter import IfHpoFilter
>>> mtc_filter = IfHpoFilter.default_filter(hpo)
Note
See the MT filters: Choosing which terms to test section for info regarding other phenotype MT filters.
then a statistical test (e.g. Fisher Exact test):
>>> from gpsea.analysis.pcats.stats import FisherExactTest
>>> count_statistic = FisherExactTest()
Note
See the gpsea.analysis.pcats.stats module for the available multiple testing procedures
(TL;DR, just Fisher Exact test at this time).
and we finalize the setup by choosing a MTC procedure (e.g. fdr_bh for Benjamini-Hochberg) along with the MTC alpha:
>>> mtc_correction = 'fdr_bh'
>>> mtc_alpha = 0.05
Note
See the Multiple-testing correction procedures section for a list of available MTC procedure codes.
The final HpoTermAnalysis is created as:
>>> from gpsea.analysis.pcats import HpoTermAnalysis
>>> analysis = HpoTermAnalysis(
... count_statistic=count_statistic,
... mtc_filter=mtc_filter,
... mtc_correction='fdr_bh',
... mtc_alpha=0.05,
... )
The analysis is identical to the one configured in the Default analysis section.
Analysis
We can now test associations between the genotype classes and the HPO terms:
>>> result = analysis.compare_genotype_vs_phenotypes(
... cohort=cohort,
... gt_clf=gt_clf,
... pheno_clfs=pheno_clfs,
... )
>>> len(result.phenotypes)
369
>>> result.total_tests
32
We tested the cohort for association between the genotype classes (gt_clf)
and HPO terms (pheno_clfs).
Thanks to phenotype MT filter, we only tested 32 out of 369 terms.
The MT filter report shows the filtering details:
>>> from gpsea.view import MtcStatsViewer
>>> mtc_viewer = MtcStatsViewer()
>>> mtc_report = mtc_viewer.process(result)
>>> mtc_report
Phenotype testing report
- Phenotype MTC filter: Independent filtering HPO filter
- Multiple testing correction: fdr_bh
| Reason | Count |
|---|---|
| Skip terms if all counts are identical to counts for a child term | 1 |
| Skipping "general" level terms | 48 |
| Skipping terms that are rare on the cohort level (in less than 40% of the cohort members) | 288 |
Genotype phenotype associations
Last, let’s explore the associations.
GPSEA displays the associations between genotypes and HPO terms in a table, one HPO term per row. The rows are ordered by the corrected p value and nominal p value in descending order.
>>> from gpsea.view import summarize_hpo_analysis
>>> summary_df = summarize_hpo_analysis(hpo, result)
>>> summary_df
Frameshift |
Other |
Corrected p values |
p values |
|
|---|---|---|---|---|
Ventricular septal defect [HP:0001629] |
19/19 (100%) |
42/71 (59%) |
0.007741654443787785 |
0.00024192670136836828 |
Absent thumb [HP:0009777] |
14/31 (45%) |
18/100 (18%) |
0.05941874425631884 |
0.0037136715160199273 |
Secundum atrial septal defect [HP:0001684] |
4/22 (18%) |
23/55 (42%) |
0.5421253568748916 |
0.06544319142266644 |
Triphalangeal thumb [HP:0001199] |
13/32 (41%) |
23/99 (23%) |
0.5421253568748916 |
0.06932119159387057 |
Muscular ventricular septal defect [HP:0011623] |
6/25 (24%) |
8/84 (10%) |
0.5421253568748916 |
0.08470708701170182 |
Amelia involving the upper limbs [HP:0009812] |
1/37 (3%) |
0/114 (0%) |
1.0 |
0.24503311258278143 |
Patent foramen ovale [HP:0001655] |
0/36 (0%) |
4/69 (6%) |
1.0 |
0.29623386322415446 |
Perimembranous ventricular septal defect [HP:0011682] |
3/25 (12%) |
6/84 (7%) |
1.0 |
0.4256518230307461 |
Short thumb [HP:0009778] |
8/30 (27%) |
25/69 (36%) |
1.0 |
0.4870099714553749 |
Absent radius [HP:0003974] |
6/25 (24%) |
9/43 (21%) |
1.0 |
0.7703831604944444 |
Short 1st metacarpal [HP:0010034] |
0/22 (0%) |
1/45 (2%) |
1.0 |
1.0 |
Common atrium [HP:0011565] |
0/38 (0%) |
1/115 (1%) |
1.0 |
1.0 |
Unroofed coronary sinus [HP:0031297] |
0/38 (0%) |
1/117 (1%) |
1.0 |
1.0 |
Abnormal long bone morphology [HP:0011314] |
13/13 (100%) |
50/50 (100%) |
1.0 |
1.0 |
Aplasia/Hypoplasia of fingers [HP:0006265] |
19/19 (100%) |
44/44 (100%) |
1.0 |
1.0 |
Aplasia/hypoplasia involving bones of the hand [HP:0005927] |
19/19 (100%) |
44/44 (100%) |
1.0 |
1.0 |
Upper limb phocomelia [HP:0009813] |
2/37 (5%) |
8/116 (7%) |
1.0 |
1.0 |
Atrial septal defect [HP:0001631] |
20/20 (100%) |
63/65 (97%) |
1.0 |
1.0 |
Abnormal atrial septum morphology [HP:0011994] |
20/20 (100%) |
64/64 (100%) |
1.0 |
1.0 |
Abnormal cardiac atrium morphology [HP:0005120] |
20/20 (100%) |
64/64 (100%) |
1.0 |
1.0 |
Abnormal hand morphology [HP:0005922] |
20/20 (100%) |
75/75 (100%) |
1.0 |
1.0 |
Aplasia/hypoplasia of the extremities [HP:0009815] |
22/22 (100%) |
78/78 (100%) |
1.0 |
1.0 |
Aplasia/hypoplasia involving bones of the upper limbs [HP:0006496] |
22/22 (100%) |
78/78 (100%) |
1.0 |
1.0 |
Aplasia/hypoplasia involving bones of the extremities [HP:0045060] |
22/22 (100%) |
78/78 (100%) |
1.0 |
1.0 |
Aplasia/hypoplasia involving the skeleton [HP:0009115] |
23/23 (100%) |
80/80 (100%) |
1.0 |
1.0 |
Abnormal cardiac septum morphology [HP:0001671] |
28/28 (100%) |
89/89 (100%) |
1.0 |
1.0 |
Complete atrioventricular canal defect [HP:0001674] |
3/36 (8%) |
6/67 (9%) |
1.0 |
1.0 |
Abnormal thumb morphology [HP:0001172] |
31/31 (100%) |
58/58 (100%) |
1.0 |
1.0 |
Abnormal finger morphology [HP:0001167] |
31/31 (100%) |
64/64 (100%) |
1.0 |
1.0 |
Abnormal digit morphology [HP:0011297] |
33/33 (100%) |
67/67 (100%) |
1.0 |
1.0 |
Abnormal appendicular skeleton morphology [HP:0011844] |
34/34 (100%) |
93/93 (100%) |
1.0 |
1.0 |
Hypoplasia of the radius [HP:0002984] |
6/14 (43%) |
34/75 (45%) |
1.0 |
1.0 |
The table shows that several HPO terms are significantly associated with presence of a heterozygous (Frameshift) frameshift variant in TBX5. For example, Ventricular septal defect was observed in 42/71 (59%) patients with no frameshift allele (Other) but it was observed in 19/19 (100%) patients with a frameshift allele (Frameshift). Fisher exact test computed a p value of ~0.000242 and the p value corrected by Benjamini-Hochberg procedure is ~0.00774.